Fragment Analyzer interpretation
Quality metrics scoring with ProSize
ProSize is the Agilent data analysis software that identifies and analyzes nucleic acid fragments and smears. ProSize software automatically calculates fragment size and quantification displaying the data in multiple formats. To interpret your results, we encourage you to consult this guide provided by Agilent.
Electropherogram interpretation tips
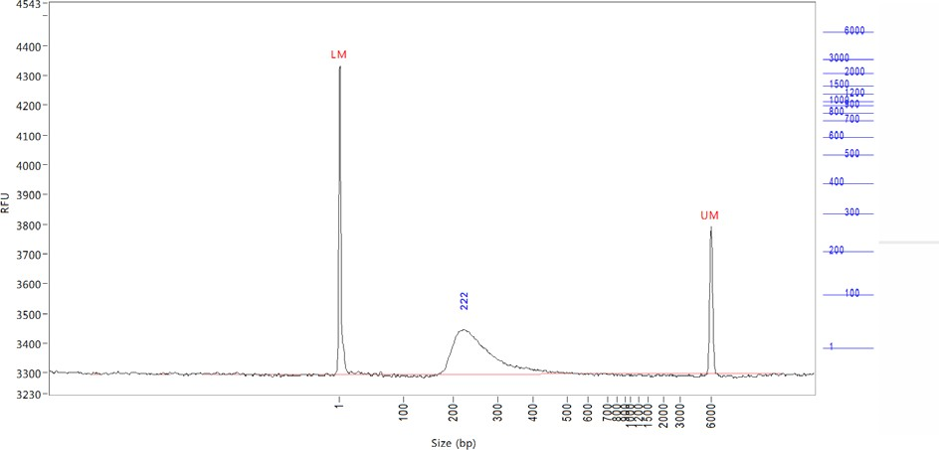
Clean Illumina library
A typical library has fragment size in the range of 200-1000 base pairs (bp). That is where you should see most of your signal. The lower marker (LM) at 1 bp and upper marker (UM) at 6000 bp are internal markers that are added to ensure proper sizing of your sample. They are added by the Genomics Facility during the loading process, and are not part of you sample.
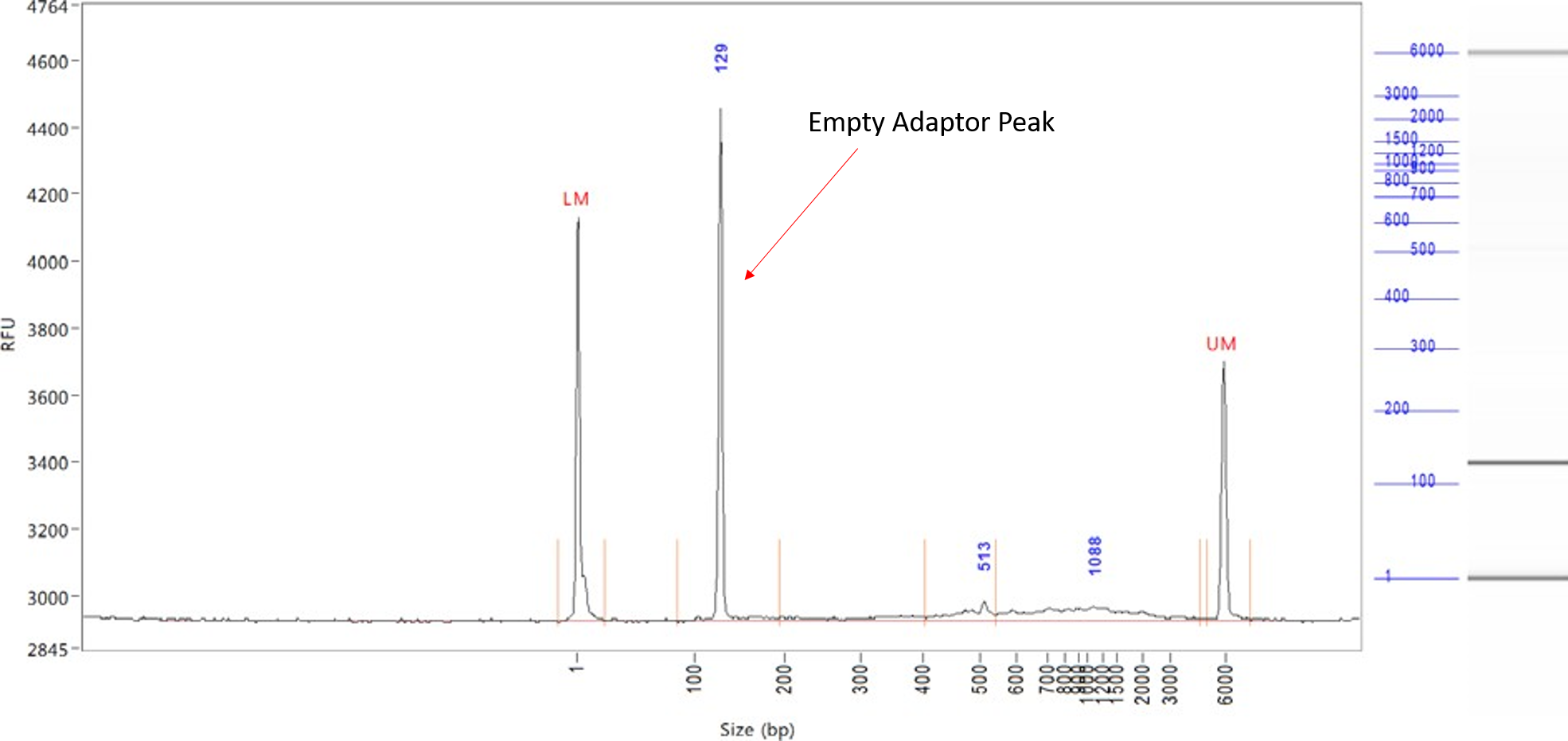
Empty adaptors library
If your library has empty adaptors (primer dimers), they will appear at ~130 bp. Empty adaptors will bind to the flowcell and sequence, and will limit the number of usable reads from your run. We recommend doing an additional clean-up on the sample in order to increase the quality of your sequencing results.
Free floating primers
Another type of contamination is by primers. Illumina’s standard primers are ~65 bp in length. They may also cause problems when sequencing if they are prominent in your sample. They can bind to the flowcell and prevent proper cluster generation, and will decrease the total yield of your sample. We recommend doing an additional clean-up on the sample in order to increase the quality of your sequencing results.
As a rule of thumb, we recommend that empty adaptors do not exceed 20% of the molarity of the sample.